Calculus - Week 3
Contents
Calculus - Week 3#
[1]:
import math
import re
import warnings
import matplotlib.pyplot as plt
import numpy as np
import plotly.graph_objects as go
import plotly.io as pio
import sympy as sp
from IPython.core.getipython import get_ipython
from IPython.display import display, HTML, Math
from matplotlib.animation import FuncAnimation
from scipy.interpolate import interp1d
plt.style.use("seaborn-v0_8-whitegrid")
pio.renderers.default = "plotly_mimetype+notebook"
Optimizing neural networks#
Single-neuron network with linear activation and Mean Squared Error (MSE) loss function#
Let the linear model \(Z = wX + b\), where
\(X\) is a \((k, m)\) matrix of \(k\) features for \(m\) samples,
\(w\) is a \((1, k)\) matrix (row vector) containing \(k\) weights,
\(b\) is a \((1, 1)\) matrix (scalar) containing 1 bias, such that
\(Z = \begin{bmatrix}w_1&&w_2&&\dots w_k\end{bmatrix} \begin{bmatrix}x_{11}&&x_{12}&&\dots&&x_{1m}\\x_{21}&&x_{22}&&\dots&&x_{2m}\\\vdots&&\vdots&&\ddots&&\vdots\\x_{k1}&&x_{k2}&&\dots&&x_{km} \end{bmatrix} + \begin{bmatrix}b\end{bmatrix}\)
\(Z\) is a \((1, m)\) matrix.
Let \(Y\) a \((1, m)\) matrix (row vector) containing the labels of \(m\) samples, such that
\(Y = \begin{bmatrix}y_1&&y_2&&\dots&&y_m\end{bmatrix}\)
[2]:
m = 40
k = 2
w = np.array([[0.37, 0.57]])
b = np.array([[0.1]])
rng = np.random.default_rng(4)
X = rng.standard_normal((k, m))
Y = w @ X + b + rng.normal(size=(1, m))
scatter = go.Scatter3d(
z=Y.squeeze(),
x=X[0],
y=X[1],
mode="markers",
marker=dict(color="#1f77b4", size=5),
name="data",
)
fig = go.Figure(scatter)
fig.update_layout(
title="Regression data",
autosize=False,
width=600,
height=600,
scene_aspectmode="cube",
margin=dict(l=10, r=10, b=10, t=30),
scene=dict(
xaxis=dict(title="x1", range=[-2.5, 2.5]),
yaxis=dict(title="x2", range=[-2.5, 2.5]),
zaxis_title="y",
camera_eye=dict(x=1.2, y=-1.8, z=0.5),
),
)
fig.show()
The predictions \(\hat{Y}\) are the result of passing \(Z\) to a linear activation function, so that \(\hat{Y} = I(Z)\).
[3]:
def linear(x, m):
return x * m
def init_neuron_params(k):
w = rng.uniform(size=(1, k)) * 0.5
b = np.zeros((1, 1))
return {"w": w, "b": b}
xx1, xx2 = np.meshgrid(np.linspace(-2.5, 2.5, 100), np.linspace(-2.5, 2.5, 100))
w, b = init_neuron_params(k).values()
random_model_plane = go.Surface(
z=linear(w[0, 0] * xx1 + w[0, 1] * xx2 + b, m=1),
x=xx1,
y=xx2,
colorscale=[[0, "#FF8920"], [1, "#FF8920"]],
showscale=False,
opacity=0.5,
name="init params",
)
fig.add_trace(random_model_plane)
fig.update_layout(title="Random model")
fig.show()
For a single sample the squared loss is
\(\ell(w, b, y_i) = (y_i - \hat{y}_i)^2 = (y_i - wx_i - b)^2\)
For the whole sample the mean squared loss is
\(\mathcal{L}(w, b, Y) = \cfrac{1}{m}\sum \limits_{i=1}^{m} \ell(w, b, y_i) = \cfrac{1}{m}\sum \limits_{i=1}^{m} (y_i - wx_i - b)^2\)
So that we don’t have a lingering 2 in the partial derivatives, we can rescale it by 0.5
\(\mathcal{L}(w, b, Y) = \cfrac{1}{2m}\sum \limits_{i=1}^{m} (y_i - wx_i - b)^2\)
[4]:
def neuron_output(X, params, activation, *args):
w = params["w"]
b = params["b"]
Z = w @ X + b
Y_hat = activation(Z, *args)
return Y_hat
def compute_mse_loss(Y, Y_hat):
m = Y_hat.shape[1]
return np.sum((Y - Y_hat) ** 2) / (2 * m)
params = init_neuron_params(k)
Y_hat = neuron_output(X, params, linear, 1)
print(f"MSE loss of random model: {compute_mse_loss(Y, Y_hat):.2f}")
MSE loss of random model: 0.49
The partial derivatives of the loss function are
\(\cfrac{\partial \mathcal{L}}{\partial w} = \cfrac{\partial \mathcal{L}}{\partial \hat{Y}}\cfrac{\partial \hat{Y}}{\partial w}\)
\(\cfrac{\partial \mathcal{L}}{\partial b} = \cfrac{\partial \mathcal{L}}{\partial \hat{Y}}\cfrac{\partial \hat{Y}}{\partial b}\)
Let’s calculate \(\cfrac{\partial \mathcal{L}}{\partial \hat{Y}}\), \(\cfrac{\partial \hat{Y}}{\partial w}\) and \(\cfrac{\partial \mathcal{L}}{\partial \hat{Y}}\)
\(\cfrac{\partial \mathcal{L}}{\partial \hat{Y}} = \cfrac{\partial}{\partial \hat{Y}} \cfrac{1}{m}\sum \limits_{i=1}^{m} (Y - \hat{Y})^2 = \cfrac{1}{m}\sum \limits_{i=1}^{m} 2(Y - \hat{Y})(- 1) = -\cfrac{1}{m}\sum \limits_{i=1}^{m}(Y - \hat{Y})\)
\(\cfrac{\partial \hat{Y}}{\partial w} = \cfrac{\partial}{\partial w} wX + b = X\)
\(\cfrac{\partial \hat{Y}}{\partial b} = wX + b = X = 1\)
Let’s put it all together
\(\cfrac{\partial \mathcal{L}}{\partial w} = -\cfrac{1}{m}\sum \limits_{i=1}^{m} (Y - \hat{Y}) \cdot X^T\)
\(\cfrac{\partial \mathcal{L}}{\partial b} = -\cfrac{1}{m}\sum \limits_{i=1}^{m} (Y - \hat{Y})\)
🔑 \(\cfrac{\partial \mathcal{L}}{\partial w}\) contains all the partial derivative wrt to each of the \(k\) elements of \(w\); it’s a \((k, m)\) matrix, because the dot product is between a matrix of shape \((1, m)\) and \(X^T\) which is \((m, k)\).
[5]:
def compute_grads(X, Y, Y_hat):
m = Y_hat.shape[1]
dw = -1 / m * np.dot(Y - Y_hat, X.T) # (1, k)
db = -1 / m * np.sum(Y - Y_hat, axis=1, keepdims=True) # (1, 1)
return {"w": dw, "b": db}
def update_params(params, grads, learning_rate=0.1):
params = params.copy()
for k in grads.keys():
params[k] = params[k] - learning_rate * grads[k]
return params
params = init_neuron_params(k)
Y_hat = neuron_output(X, params, linear, 1)
print(f"MSE loss before update: {compute_mse_loss(Y, Y_hat):.2f}")
grads = compute_grads(X, Y, Y_hat)
params = update_params(params, grads)
Y_hat = neuron_output(X, params, linear, 1)
print(f"MSE loss after update: {compute_mse_loss(Y, Y_hat):.2f}")
MSE loss before update: 0.50
MSE loss after update: 0.48
Let’s find the best parameters with gradient descent.
[6]:
params = init_neuron_params(k)
Y_hat = neuron_output(X, params, linear, 1)
loss = compute_mse_loss(Y, Y_hat)
print(f"Iter 0 - MSE loss={loss:.6f}")
for i in range(1, 50 + 1):
grads = compute_grads(X, Y, Y_hat)
params = update_params(params, grads)
Y_hat = neuron_output(X, params, linear, 1)
loss_new = compute_mse_loss(Y, Y_hat)
if loss - loss_new <= 1e-4:
print(f"Iter {i} - MSE loss={loss:.6f}")
print("The algorithm has converged")
break
loss = loss_new
if i % 5 == 0:
print(f"Iter {i} - MSE loss={loss:.6f}")
Iter 0 - MSE loss=0.439887
Iter 5 - MSE loss=0.420053
Iter 10 - MSE loss=0.412665
Iter 15 - MSE loss=0.409886
Iter 20 - MSE loss=0.408831
Iter 22 - MSE loss=0.408717
The algorithm has converged
Let’s visualize the final model.
[7]:
w, b = params.values()
final_model_plane = go.Surface(
z=w[0, 0] * xx1 + w[0, 1] * xx2 + b,
x=xx1,
y=xx2,
colorscale=[[0, "#2ca02c"], [1, "#2ca02c"]],
showscale=False,
opacity=0.5,
name="final params",
)
fig.add_trace(final_model_plane)
fig.data[1].visible = False
fig.update_layout(title="Final model")
fig.show()
Single-neuron network with sigmoid activation and Log loss function#
As before, let the linear model \(Z = wX + b\) and \(Y\) a row vector containing the labels of \(m\) samples.
[8]:
m = 40
k = 2
neg_centroid = [-1, -1]
pos_centroid = [1, 1]
rng = np.random.default_rng(1)
X = np.r_[
rng.standard_normal((m // 2, k)) + neg_centroid,
rng.standard_normal((m // 2, k)) + pos_centroid,
].T
Y = np.array([[0] * (m // 2) + [1] * (m // 2)])
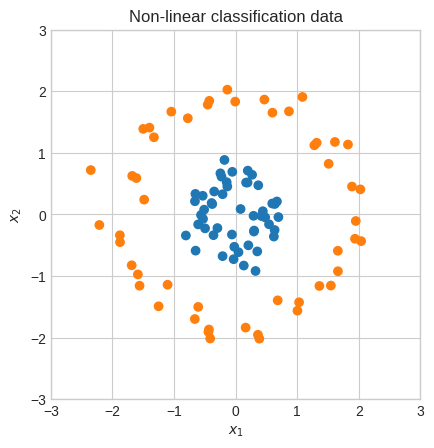
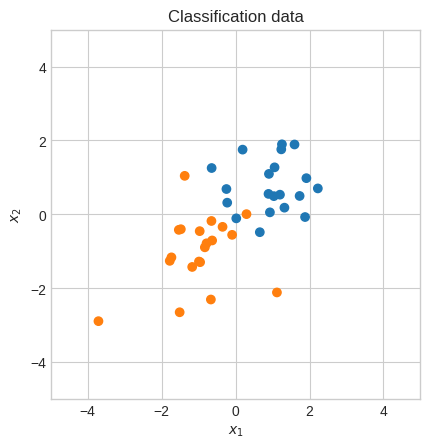
plt.scatter(X[0], X[1], c=np.where(Y.squeeze() == 0, "tab:orange", "tab:blue"))
plt.gca().set_aspect("equal")
plt.xlabel("$x_1$")
plt.ylabel("$x_2$")
plt.xlim(-5, 5)
plt.ylim(-5, 5)
plt.title("Classification data")
plt.show()
This time, though, the predictions \(\hat{Y}\) are the result of passing \(Z\) to a sigmoid function, so that \(\hat{Y} = \sigma(Z)\).
\(\sigma(Z) = \cfrac{1}{1+e^{-Z}}\)
To visualize the sigmoid function let’s add another axis.
[9]:
neg_scatter = go.Scatter3d(
z=np.full(int(m / 2), 0),
x=X[0, : int(m / 2)],
y=X[1, : int(m / 2)],
mode="markers",
marker=dict(color="#ff7f0e", size=5),
name="negative class",
)
pos_scatter = go.Scatter3d(
z=np.full(int(m / 2), 1),
x=X[0, int(m / 2) :],
y=X[1, int(m / 2) :],
mode="markers",
marker=dict(color="#1f77b4", size=5),
name="positive class",
)
fig = go.Figure([pos_scatter, neg_scatter])
fig.update_layout(
title="Classification data",
autosize=False,
width=600,
height=600,
margin=dict(l=10, r=10, b=10, t=30),
scene=dict(
xaxis=dict(title="x1", range=[-5, 5]),
yaxis=dict(title="x2", range=[-5, 5]),
zaxis_title="y",
camera_eye=dict(x=0, y=0.3, z=2.5),
camera_up=dict(x=0, y=np.sin(np.pi), z=0),
),
)
Now, let’s plot the predictions of a randomly initialized model.
[10]:
def sigmoid(x):
return 1 / (1 + np.exp(-x))
xx1, xx2 = np.meshgrid(np.linspace(-5, 5, 100), np.linspace(-5, 5, 100))
w, b = init_neuron_params(k).values()
random_model_plane = go.Surface(
z=sigmoid(w[0, 0] * xx1 + w[0, 1] * xx2 + b),
x=xx1,
y=xx2,
colorscale=[[0, "#ff7f0e"], [1, "#1f77b4"]],
showscale=False,
opacity=0.5,
name="init params",
)
fig.add_trace(random_model_plane)
fig.update_layout(
title="Random model",
)
fig.show()
It turns out that the output of the sigmoid activation is the probability \(p\) of a sample belonging to the positive class, which implies that \((1-p)\) is the probability of a sample belonging to the negative class.
Intuitively, a loss function will be small when \(p_i\) is close to 1.0 and \(y_i = 1\) and when \(p_i\) is close to 0.0 and \(y_i = 0\).
For a single sample this loss function might look like this
\(\ell(w, b, y_i) = -p_i^{y_i}(1-p_i)^{1-y_i}\)
For the whole sample the loss would be
\(\mathcal{L}(w, b, Y) = -\prod \limits_{i=1}^{m} p_i^{y_i}(1-p_i)^{1-y_i}\)
In Univariate optimization (week 1) we’ve seen how it’s easier to calculate the derivative of the logarithm of the PMF of the Binomial Distribution.
We can do the same here to obtain a more manageable loss function.
\(\mathcal{L}(w, b, Y) = -\sum \limits_{i=1}^{m} y_i \ln p_i + (1-y_i) \ln (1-p_i)\)
It turns out it’s standard practice to minimize a function and to average the loss over the sample (to manage the scale of the loss for large datasets), so we’ll use this instead:
\(\mathcal{L}(w, b, Y) = -\cfrac{1}{m} \sum \limits_{i=1}^{m} y_i \ln p_i + (1-y_i) \ln (1-p_i)\)
Finally, let’s substitute back \(\hat{y}_i\) for \(p_i\).
\(\mathcal{L}(w, b, Y) = -\cfrac{1}{m} \sum \limits_{i=1}^{m} y_i \ln \hat{y}_i + (1-y_i) \ln (1-\hat{y}_i)\)
Recall that \(\hat{y}_i = \sigma(z_i)\).
[11]:
def compute_log_loss(Y, Y_hat):
m = Y_hat.shape[1]
loss = (-1 / m) * (
np.dot(Y, np.log(Y_hat).T) + np.dot((1 - Y), np.log(1 - Y_hat).T)
)
return loss.squeeze()
params = init_neuron_params(k)
Y_hat = neuron_output(X, params, sigmoid)
print(f"Log loss of random model: {compute_log_loss(Y, Y_hat):.2f}")
Log loss of random model: 0.50
The partial derivatives of the loss function are
\(\cfrac{\partial \mathcal{L}}{\partial w} = \cfrac{\partial \mathcal{L}}{\partial \hat{Y}}\cfrac{\partial \hat{Y}}{\partial w}\)
\(\cfrac{\partial \mathcal{L}}{\partial b} = \cfrac{\partial \mathcal{L}}{\partial \hat{Y}}\cfrac{\partial \hat{Y}}{\partial b}\)
Because we have an activation function around \(Z\), we have another chain rule to apply.
For the MSE loss, the activation was an identity function so it didn’t pose the same challenge.
\(\cfrac{\partial \mathcal{L}}{\partial w} = \cfrac{\partial \mathcal{L}}{\partial \sigma(Z)}\cfrac{\partial \sigma(Z)}{\partial w} = \cfrac{\partial \mathcal{L}}{\partial \sigma(Z)}\cfrac{\partial \sigma(Z)}{\partial Z}\cfrac{\partial Z}{\partial w}\)
\(\cfrac{\partial \mathcal{L}}{\partial b} = \cfrac{\partial \mathcal{L}}{\partial \sigma(Z)}\cfrac{\partial \sigma(Z)}{\partial b} = \cfrac{\partial \mathcal{L}}{\partial \sigma(Z)}\cfrac{\partial \sigma(Z)}{\partial Z}\cfrac{\partial Z}{\partial b}\)
Let’s calculate \(\cfrac{\partial \mathcal{L}}{\partial \sigma(Z)}\), \(\cfrac{\partial \sigma(Z)}{\partial Z}\), \(\cfrac{\partial Z}{\partial w}\) and \(\cfrac{\partial Z}{\partial b}\)
Calculation of partial derivative of Log loss wrt sigmoid activation#
\(\cfrac{\partial \mathcal{L}}{\partial \sigma(Z)} = \cfrac{\partial}{\partial \sigma(Z)} -\cfrac{1}{m} \sum \limits_{i=1}^{m} Y\ln\sigma(Z)+(Y-1)\ln(1-\sigma(Z))\)
\(= -\cfrac{1}{m} \sum \limits_{i=1}^{m} \cfrac{Y}{\sigma(Z)}+\cfrac{Y-1}{1-\sigma(Z)}\)
\(= -\cfrac{1}{m} \sum \limits_{i=1}^{m} \cfrac{Y(1-\sigma(Z)) + (Y-1)\sigma(Z)}{\sigma(Z)(1-\sigma(Z))}\)
\(= -\cfrac{1}{m} \sum \limits_{i=1}^{m} \cfrac{Y -\sigma(Z)}{\sigma(Z)(1-\sigma(Z))}\)
Calculation of the derivative of the sigmoid function#
\(\cfrac{\partial \sigma(Z)}{\partial Z} = \cfrac{\partial}{\partial Z} \cfrac{1}{1+e^{-Z}}\)
\(= (1+e^{-Z})^{-1}\)
\(= -(1+e^{-Z})^{-2}(-e^{-Z})\)
\(= (1+e^{-Z})^{-2}e^{-Z}\)
\(= \cfrac{e^{-Z}}{(1+e^{-Z})^2}\)
\(= \cfrac{1 +e^{-Z} - 1}{(1+e^{-Z})^2}\)
\(= \cfrac{1 +e^{-Z}}{(1+e^{-Z})^2}-\cfrac{1}{(1+e^{-Z})^2}\)
\(= \cfrac{1}{1+e^{-Z}}-\cfrac{1}{(1+e^{-Z})^2}\)
Because the factor of \(x-x^2\) is \(x(1-x)\) we can factor it to
\(= \cfrac{1}{1+e^{-Z}}\left(1 - \cfrac{1}{1+e^{-Z}} \right)\)
\(= \sigma(Z)(1 - \sigma(Z))\)
The other two partial derivatives are the same as the MSE loss’.
\(\cfrac{\partial Z}{\partial w} = \cfrac{\partial}{\partial w} wX+b = X\)
\(\cfrac{\partial Z}{\partial b} = \cfrac{\partial}{\partial b} wX+b = 1\)
Let’s put it all together
\(\cfrac{\partial \mathcal{L}}{\partial w} = -\cfrac{1}{m} \sum \limits_{i=1}^{m} \cfrac{Y -\sigma(Z)}{\sigma(Z)(1-\sigma(Z))} \sigma(Z)(1 - \sigma(Z)) X^T = -\cfrac{1}{m} \sum \limits_{i=1}^{m} (Y -\sigma(Z))X^T = -\cfrac{1}{m} \sum \limits_{i=1}^{m} (Y - \hat{Y}) X^T\)
\(\cfrac{\partial \mathcal{L}}{\partial b} = -\cfrac{1}{m} \sum \limits_{i=1}^{m} \cfrac{Y -\sigma(Z)}{\sigma(Z)(1-\sigma(Z))} \sigma(Z)(1 - \sigma(Z)) = -\cfrac{1}{m} \sum \limits_{i=1}^{m} Y - \hat{Y}\)
🔑 The partial derivatives of the Log loss \(\cfrac{\partial \mathcal{L}}{\partial w}\) and \(\cfrac{\partial \mathcal{L}}{\partial b}\) are the same as the MSE loss’.
[12]:
params = init_neuron_params(k)
Y_hat = neuron_output(X, params, sigmoid)
print(f"Log loss before update: {compute_log_loss(Y, Y_hat):.2f}")
grads = compute_grads(X, Y, Y_hat)
params = update_params(params, grads)
Y_hat = neuron_output(X, params, sigmoid)
print(f"Log loss after update: {compute_log_loss(Y, Y_hat):.2f}")
Log loss before update: 0.53
Log loss after update: 0.51
Let’s find the best parameters using gradient descent.
[13]:
params = init_neuron_params(k)
Y_hat = neuron_output(X, params, sigmoid)
loss = compute_log_loss(Y, Y_hat)
print(f"Iter 0 - Log loss={loss:.6f}")
for i in range(1, 500 + 1):
grads = compute_grads(X, Y, Y_hat)
params = update_params(params, grads)
Y_hat = neuron_output(X, params, sigmoid)
loss_new = compute_log_loss(Y, Y_hat)
if loss - loss_new <= 1e-4:
print(f"Iter {i} - Log loss={loss:.6f}")
print("The algorithm has converged")
break
loss = loss_new
if i % 100 == 0:
print(f"Iter {i} - Log loss={loss:.6f}")
Iter 0 - Log loss=0.582076
Iter 100 - Log loss=0.182397
Iter 200 - Log loss=0.148754
Iter 300 - Log loss=0.134595
Iter 306 - Log loss=0.134088
The algorithm has converged
Let’s visualize the final model.
[14]:
w, b = params.values()
final_model_plane = go.Surface(
z=sigmoid(w[0, 0] * xx1 + w[0, 1] * xx2 + b),
x=xx1,
y=xx2,
colorscale=[[0, "#ff7f0e"], [1, "#1f77b4"]],
showscale=False,
opacity=0.5,
name="final params",
)
fig.add_trace(final_model_plane)
fig.data[2].visible = False
fig.update_layout(title="Final model")
fig.show()
Newton-Raphson method#
Newton-Raphson method with one variable#
🔑 Newton-Raphson method is an algorithm to find the zero of a function
It can be used to optimize a function \(f(x)\) by finding the zero of \(f'(x)\).
The method works by starting from a random point \(x_0\) on \(f'(x)\), finding the tangent at \(f'(x_0)\) and finding where the tangent crosses the x-axis.
The idea is that this new point is closer than \(x_0\) to a minimum or maximum.
[24]:
def fixup_animation_js(html_animation):
html_animation = html_animation.replace(
'<div class="anim-controls">',
'<div class="anim-controls" style="display:none">',
)
animation_id = re.findall(r"onclick=\"(.*)\.", html_animation)[0]
img_id = re.findall(r"<img id=\"(.*)\"", html_animation)[0]
html_animation += f"""
<script language="javascript">
setupAnimationIntersectionObserver('{animation_id}', '{img_id}');
</script>
"""
return html_animation
def _update(frame):
global fscat, dscat, dtang
fscat.remove()
dscat.remove()
dtang.remove()
fscat = ax.scatter(xs[frame], f(xs[frame]), color="tab:blue")
dscat = ax.scatter(xs[frame], df(xs[frame]), color="tab:orange")
(dtang,) = ax.plot(
xx,
ddf(xs[frame]) * (xx - xs[frame]) + df(xs[frame]),
"--",
color="tab:orange",
alpha=0.5,
)
x = sp.symbols("x")
f = sp.exp(x) - sp.log(x)
df = sp.lambdify(x, sp.diff(f, x, 1))
ddf = sp.lambdify(x, sp.diff(f, x, 2))
f = sp.lambdify(x, f)
xs = [1.75]
for _ in range(4):
xs.append(xs[-1] - df(xs[-1]) / ddf(xs[-1]))
fig, ax = plt.subplots()
xx = np.linspace(1e-3, 2)
ax.plot(xx, f(xx), color="tab:blue", label="$f(x)$")
ax.plot(xx, df(xx), color="tab:orange", label="$f'(x)$")
fscat = ax.scatter([], [])
dscat = ax.scatter([], [])
(dtang,) = ax.plot([], [])
ax.set_ylim(-2, 11)
ax.set_xlim(0, 2)
ax.spines[["left", "bottom"]].set_position(("data", 0))
ax.text(1, 0, "$x$", transform=ax.get_yaxis_transform())
ax.text(0, 1.02, "$f(x)$", transform=ax.get_xaxis_transform())
plt.legend(loc="upper right")
plt.title("Minimazation of $f(x)$ using Newton's method")
ani = FuncAnimation(fig=fig, func=_update, frames=4, interval=1000)
html_animation = ani.to_jshtml(default_mode="loop")
if "runtime" not in get_ipython().config.IPKernelApp.connection_file:
html_animation = fixup_animation_js(html_animation)
display(HTML(html_animation))
plt.close()
The equation of a line that passes for \(\left(x_0, f(x_0)\right)\) is the point-slope form
\(f(x) - f(x_0) = m(x-x_0)\)
\(f(x) = m(x - x_0) + f(x_0)\)
We substitute \(f'(x)\) for \(f(x)\), \(f'(x_0)\) for \(f(x_0)\), and the derivative of \(f'(x)\) at \(x_0\) (ie the second derivative \(f''(x_0)\)) for \(m\) and we obtain
\(f'(x) = f''(x_0)(x - x_0) + f'(x_0)\)
Now we want to find the \(x\) such that \(f'(x) = 0\), that is the point where the tangent crosses the x-axis.
\(0 = f''(x_0)(x - x_0) + f'(x_0)\)
\(-f''(x_0)(x - x_0) = f'(x_0)\)
\(-(x - x_0) = \cfrac{f'(x_0)}{f''(x_0)}\)
\(-x + x_0 = \cfrac{f'(x_0)}{f''(x_0)}\)
\(-x = - x_0 + \cfrac{f'(x_0)}{f''(x_0)}\)
\(x = x_0 - \cfrac{f'(x_0)}{f''(x_0)}\)
The pseudo-code might be looking something along these lines
\(\begin{array}{l} \textbf{Algorithm: } \text{Newton's Method for Univariate Optimization} \\ \textbf{Input: } \text{function to minimize } \mathcal{f}, \text{initial guess } x_0, \text{number of iterations } K, \text{convergence } \epsilon \\ \textbf{Output: } x \\ 1 : x \gets x_0 \\ 2 : \text{for k = 1 to K do} \\ 3 : \quad x_{new} = x - f'(x) / f''(x) \\ 4 : \quad \text{if } \|f'(x_{new}) / f''(x_{new})\| < \epsilon \text{ then} \\ 5 : \quad \quad \text{return } x \\ 6 : \quad \text {end if} \\ 7 : \quad x \gets x_{new} \\ 8 : \text{return } x \\ \end{array}\)
Another way to find \(x\) such that \(f'(x) = 0\) is through the rise over run equation.
\(f''(x_0) = \cfrac{f'(x_0)}{x_0 - x}\)
\(\cfrac{1}{f''(x_0)} = \cfrac{x_0 - x}{f'(x_0)}\)
\(\cfrac{f'(x_0)}{f''(x_0)} = x_0 - x\)
\(\cfrac{f'(x_0)}{f''(x_0)} - x_0 = - x\)
\(x_0 - \cfrac{f'(x_0)}{f''(x_0)} = x\)
A third way to find \(x\) such that \(f'(x) = 0\) is through expressing \(f(x)\) as a quadratic approximation (Taylor series truncated at the second order).
A Taylor series of \(f(x)\) that is differentiable at \(x_0\) is
\(f(x)=\sum_{n=0}^\infty\cfrac{f^{(n)}(x_0)(x-x_0)^n}{n!}\)
Before proceeding let’s take a look at what a Taylor series is.
[25]:
def _update(order):
global taylor
taylor.remove()
series = np.zeros((order, 50))
for n in range(1, order + 1):
series[n - 1] = (
(1 / math.factorial(n))
* sp.diff(expr, x, n).evalf(subs={x: 0})
* (xx**n)
)
(taylor,) = ax.plot(xx, np.sum(series, axis=0), color="tab:orange")
x = sp.symbols("x")
expr = sp.sin(x)
sin = sp.lambdify(x, expr, "numpy")
xx = np.linspace(-10, 10)
fig, ax = plt.subplots()
ax.plot(xx, sin(xx), color="tab:blue", label="$\sin(x)$")
(taylor,) = ax.plot([], [], color="tab:orange", label="Taylor series")
ax.set_aspect("equal")
ax.set_ylim(-10, 10)
ax.set_xlim(-10, 10)
plt.legend(loc="upper right")
plt.title("Taylor series of order 1, 3, ..., 25 of sine function")
ani = FuncAnimation(fig=fig, func=_update, frames=range(1, 27, 2))
html_animation = ani.to_jshtml(default_mode="loop")
if "runtime" not in get_ipython().config.IPKernelApp.connection_file:
html_animation = fixup_animation_js(html_animation)
display(HTML(html_animation))
plt.close()
🔑 The Taylor series of a function is an infinite sum of terms (polynomials) that are expressed in terms of the function derivatives at a point.
The quadratic approximation of \(f(x)\) (Taylor series truncated at the second degree polynomial) is
\(f(x) \approx f(x_0) + f'(x_0)(x-x_0) + \cfrac{1}{2}f''(x_0)(x-x_0)^2\)
To find \(x\) such that \(f'(x) = 0\), we set the derivative of the quadratic approximation to zero.
\(\cfrac{d}{dx}f(x_0) + f'(x_0)(x-x_0) + \cfrac{1}{2}f''(x_0)(x-x_0)^2 = 0\)
\(f'(x_0) + f''(x_0)(x-x_0) = 0\)
\(f''(x_0)(x-x_0) = -f'(x_0)\)
\(x-x_0 = -\cfrac{f'(x_0)}{f''(x_0)}\)
\(x = x_0 -\cfrac{f'(x_0)}{f''(x_0)}\)
This derivation of the Newton’s method update formula from the quadratic approximation of a function has a few important implications:
At each step, Newton’s method finds the minimum of the quadratic approximation of \(f(x)\)
When \(f(x)\) is indeed quadratic, Newton’s method will find the minimum or maximum in one single step
When \(f(x)\) is not quadratic, Newton’s method will exhibit quadratic convergence near the minimum or maximum. Quadratic convergence refers to the rate at which the sequence \({x_k}\) approaches the minimum or maximum. The “error” will decrease quadratically at each step.
[26]:
def _update(frame):
global fscat, taylor, taylor_min
fscat.remove()
taylor.remove()
taylor_min.remove()
fscat = ax.scatter(xs[frame], f(xs[frame]), color="tab:blue")
(taylor,) = ax.plot(
xx,
f(xs[frame])
+ df(xs[frame]) * (xx - xs[frame])
+ (1 / 2) * ddf(xs[frame]) * (xx - xs[frame]) ** 2,
"--",
color="tab:orange",
alpha=0.5,
label=r"$f(x_0) + f'(x_0)(x-x_0) + \dfrac{1}{2}f''(x_0)(x-x_0)^2$",
)
taylor_min = ax.scatter(
xs[frame + 1],
f(xs[frame])
+ df(xs[frame]) * (xs[frame + 1] - xs[frame])
+ (1 / 2) * ddf(xs[frame]) * (xs[frame + 1] - xs[frame]) ** 2,
color="tab:orange",
)
x = sp.symbols("x")
f = sp.exp(x) - sp.log(x)
df = sp.lambdify(x, sp.diff(f, x, 1))
ddf = sp.lambdify(x, sp.diff(f, x, 2))
f = sp.lambdify(x, f)
xs = [1.75]
for _ in range(4):
xs.append(xs[-1] - df(xs[-1]) / ddf(xs[-1]))
fig, ax = plt.subplots()
xx = np.linspace(1e-3, 2)
ax.plot(xx, f(xx), color="tab:blue", label="$f(x)$")
fscat = ax.scatter([], [])
(taylor,) = ax.plot(
[],
[],
color="tab:orange",
alpha=0.5,
label=r"$f(x_0) + f'(x_0)(x-x_0) + \dfrac{1}{2}f''(x_0)(x-x_0)^2$",
)
taylor_min = ax.scatter([], [])
ax.set_ylim(-2, 11)
ax.set_xlim(0, 2)
ax.spines[["left", "bottom"]].set_position(("data", 0))
ax.text(1, 0, "$x$", transform=ax.get_yaxis_transform())
ax.text(0, 1.02, "$f(x)$", transform=ax.get_xaxis_transform())
plt.legend(loc="upper right")
plt.title(
"At each step, Newton's method finds\nthe minimum of the quadratic approximation of $f(x)$"
)
ani = FuncAnimation(fig=fig, func=_update, frames=4, interval=1000)
html_animation = ani.to_jshtml(default_mode="loop")
if "runtime" not in get_ipython().config.IPKernelApp.connection_file:
html_animation = fixup_animation_js(html_animation)
display(HTML(html_animation))
plt.close()
Finally, let’s inspect the algorithm.
[27]:
def univariate_newtons_method(f, symbols, initial, steps):
x = symbols
df = sp.lambdify(x, sp.diff(f, x, 1))
ddf = sp.lambdify(x, sp.diff(f, x, 2))
p = np.zeros((steps, 1))
g = np.zeros((steps, 1))
h = np.zeros((steps, 1))
step_vector = np.zeros((steps, 1))
p[0] = initial
g[0] = df(p[0])
h[0] = ddf(p[0])
step_vector[0] = df(p[0]) / ddf(p[0])
for i in range(1, steps):
p[i] = p[i - 1] - step_vector[i - 1]
g[i] = df(p[i])
h[i] = ddf(p[i])
step_vector[i] = df(p[i]) / ddf(p[i])
if np.linalg.norm(step_vector[i]) < 1e-4:
break
return p[: i + 1], g[: i + 1], h[: i + 1], step_vector[: i + 1]
x = sp.symbols("x")
f = sp.exp(x) - sp.log(x)
p, _, _, _ = univariate_newtons_method(f, symbols=x, initial=1.75, steps=10)
display(
Math(
"\\arg \\min \\limits_{x}" + sp.latex(f) + f"={p[-1].squeeze():.4f}"
)
)
print(f"Newton's method converged after {len(p)-1} steps")
Newton's method converged after 4 steps
Let’s consider the quadratic function \(f(x) = x^2\) which has a minimum at 0.
Based on what we said above we expect Newton’s method to find this minimum in one step.
[28]:
x = sp.symbols("x")
f = x**2
p, _, _, _ = univariate_newtons_method(f, symbols=x, initial=1.75, steps=10)
display(
Math(
"\\arg \\min \\limits_{x}" + sp.latex(f) + f"={p[-1].squeeze():.4f}"
)
)
print(f"Newton's method converged after {len(p)-1} steps")
Newton's method converged after 1 steps
Let’s consider the quadratic function \(f(x) = -x^2\) which has a maximum at 0.
Based on what we said above we expect Newton’s method to find this maximum in one step.
[29]:
x = sp.symbols("x")
f = -(x**2)
p, _, _, _ = univariate_newtons_method(f, symbols=x, initial=1.75, steps=10)
display(
Math(
"\\arg \\min \\limits_{x}"
+ sp.latex(f)
+ f"={p[-1].squeeze():.4f}"
)
)
print(f"Newton's method converged after {len(p)-1} steps")
Newton's method converged after 1 steps
Second derivatives#
In the previous section we introduced second derivatives without too much explanation of what they are.
After all, analytically, it’s just the derivative of a derivative.
But what do they represent and what they’re used for?
Consider this analogy.
🔑 Let \(f(t)\) be the distance, then the first derivative is velocity (instantaneous rate of change of distance wrt time), then the second derivative is acceleration (instantaneous rate of change of velocity wrt time).
[30]:
def _update(raw_frame):
global distance_point, velocity_point, acceleration_point
distance_point.remove()
velocity_point.remove()
acceleration_point.remove()
frame = int(frame_func(raw_frame))
xxf = xx[: frame + 1]
distance_line = ax.plot(
xxf, sp.lambdify(x, norm_pdf, "numpy")(xxf), c="tab:blue"
)
velocity_line = ax.plot(
xxf,
sp.lambdify(x, sp.diff(norm_pdf, x, 1), "numpy")(xxf),
c="tab:orange",
)
acceleration_line = ax.plot(
xxf,
sp.lambdify(x, sp.diff(norm_pdf, x, 2), "numpy")(xxf),
c="tab:green",
)
distance_point = ax.scatter(
xx[frame], sp.lambdify(x, norm_pdf, "numpy")(xx[frame]), color="tab:blue"
)
velocity_point = ax.scatter(
xx[frame],
sp.lambdify(x, sp.diff(norm_pdf, x, 1), "numpy")(xx[frame]),
color="tab:orange",
)
acceleration_point = ax.scatter(
xx[frame],
sp.lambdify(x, sp.diff(norm_pdf, x, 2), "numpy")(xx[frame]),
color="tab:green",
)
x = sp.symbols("x")
norm_pdf = (1 / sp.sqrt(2 * np.pi)) * sp.exp(-(x**2) / 2)
xx = np.linspace(-3, 3, 100)
fig, ax = plt.subplots()
distance_line = ax.plot([], [], c="tab:blue", label="distance")
velocity_line = ax.plot([], [], c="tab:orange", label="velocity")
acceleration_line = ax.plot([], [], c="tab:green", label="acceleration")
distance_point = ax.scatter([], [])
velocity_point = ax.scatter([], [])
acceleration_point = ax.scatter([], [])
ax.spines["bottom"].set_position(("data", 0))
plt.xlim(-3, 3)
plt.ylim(-0.5, 0.5)
plt.legend()
plt.title("Distance, velocity and acceleration analogy")
# Create velocity as a function of frame
# Takes values between 0 and 100 as arguments and returns the velocity-adjusted frame value.
# For example between 0 and 20 velocity is slow
# Frames instead of going 0, 1, 2 will go 0, 0, 1 etc.
velocity_func = sp.lambdify(x, sp.diff(norm_pdf, x, 1), "numpy")
velocity_magnitudes = np.abs(velocity_func(xx))
normalized_magnitudes = (
velocity_magnitudes / np.sum(velocity_magnitudes) * (len(xx) - 1)
)
adjusted_frame_positions = np.cumsum(normalized_magnitudes)
frame_func = interp1d(
np.arange(len(xx)),
adjusted_frame_positions,
kind="linear",
fill_value="extrapolate",
)
ani = FuncAnimation(fig=fig, func=_update, frames=len(xx))
html_animation = ani.to_jshtml(default_mode="loop")
if "runtime" not in get_ipython().config.IPKernelApp.connection_file:
html_animation = fixup_animation_js(html_animation)
display(HTML(html_animation))
plt.close()
🔑 The second derivative tells us about the curvature of a function, or in other words if a function is concave up or concave down.
When the second derivative is positive the function is concave up.
When the second derivative is negative the function is concave down.
When the second derivative is 0 the curvature of the function is undetermined.
This information in combination with the first derivative tells us about our position relative to a minimum or maximum.
\(f'(x) > 0\) and \(f''(x) > 0\): we’re on the right hand side of a concave up function (minimum to our left) (like between -3 and -1 on the graph above).
\(f'(x) > 0\) and \(f''(x) < 0\): we’re on the left hand side of a concave down function (maximum to our right) (like between -1 and 0 on the graph above).
\(f'(x) < 0\) and \(f''(x) < 0\): we’re on the right hand side of a concave down function (maximum to our left) (like between 0 and 1 on the graph above).
\(f'(x) < 0\) and \(f''(x) > 0\): we’re on the left hand side of a concave up function (minimum to our right) (like between 1 and 3 on the graph above).
Let’s see what happens when the (vanilla) Newton’s method gets to a point where the second derivative is 0.
[31]:
with warnings.catch_warnings():
warnings.filterwarnings("error")
_, _, _, _ = univariate_newtons_method(norm_pdf, symbols=x, initial=1, steps=10)
---------------------------------------------------------------------------
RuntimeWarning Traceback (most recent call last)
Cell In[31], line 3
1 with warnings.catch_warnings():
2 warnings.filterwarnings("error")
----> 3 _, _, _, _ = univariate_newtons_method(norm_pdf, symbols=x, initial=1, steps=10)
Cell In[27], line 12, in univariate_newtons_method(f, symbols, initial, steps)
10 g[0] = df(p[0])
11 h[0] = ddf(p[0])
---> 12 step_vector[0] = df(p[0]) / ddf(p[0])
13 for i in range(1, steps):
14 p[i] = p[i - 1] - step_vector[i - 1]
RuntimeWarning: divide by zero encountered in divide
Second-order partial derivatives#
For functions of more than one variable we have partial derivatives as well as second-order partial derivatives.
For example, let \(f(x, y) = 2x^2+3y^2 - xy\). We have 2 partial derivatives
\(\cfrac{\partial f}{\partial x} = 4x - y\)
\(\cfrac{\partial f}{\partial y} = 6y - x\)
and 4 second-order partial derivatives
\(\partial\cfrac{\partial f / \partial x}{\partial x} = \cfrac{\partial^2 f}{\partial x^2} = 4\)
\(\partial\cfrac{\partial f / \partial x}{\partial y} = \cfrac{\partial^2 f}{\partial x \partial y} = -1\)
\(\partial\cfrac{\partial f / \partial y}{\partial y} = \cfrac{\partial^2 f}{\partial y^2} = 6\)
\(\partial\cfrac{\partial f / \partial y}{\partial x} = \cfrac{\partial^2 f}{\partial y \partial x} = -1\)
The fact that \(\cfrac{\partial^2 f}{\partial x \partial y} = \cfrac{\partial^2 f}{\partial y \partial x}\) is not a coincidence but rather the symmetry property of partial second-order derivatives.
Hessian matrix#
If we can summarize partial derivatives into a gradient vector \(\nabla_f = \langle \cfrac{\partial f}{\partial x}, \cfrac{\partial f}{\partial y} \rangle\), it seems almost obvious that we can summarize second-order partial derivatives into a matrix.
Such matrix is called Hessian.
\(H_{f(x, y)} = \begin{bmatrix} \cfrac{\partial^2 f}{\partial x^2}&&\cfrac{\partial^2 f}{\partial x \partial y}\\ \cfrac{\partial^2 f}{\partial y \partial x}&&\cfrac{\partial^2 f}{\partial y^2} \end{bmatrix}\)
🔑 The Hessian matrix is a symmetric matrix containing the second-order partial derivatives of a function
Continuing the previous example, the Hessian matrix of \(f(x, y) = 2x^2+3y^2 - xy\) (at any point over its domain) is
\(H = \begin{bmatrix}4&&-1\\-1&&6\end{bmatrix}\)
We can also show that the original function can be expressed as
\(q(x, y) = \cfrac{1}{2}\begin{bmatrix}x&&y\end{bmatrix}\begin{bmatrix}4&&-1\\-1&&6\end{bmatrix}\begin{bmatrix}x\\y\end{bmatrix}\)
\(= \cfrac{1}{2}\begin{bmatrix}x&&y\end{bmatrix}\begin{bmatrix}4x - y\\6y - x\end{bmatrix}\)
\(= \cfrac{1}{2}(x(4x - y)+y(6y - x))\)
\(= \cfrac{1}{2}(4x^2 - xy + 6y^2 - xy)\)
\(= 2x^2 + 3y^2 - xy\)
It turns out that \(q(x, y) = x^THx\) creates a concave up function if \(H\) is a positive-definite matrix (\(x^THx>0\)) and a concave down function if H is a negative-definite matrix (\(x^THx<0\)).
How do we determine the positive definiteness of \(H\)?
With its eigenvalues.
Recall that an eigenvector is a vector \(x\) such that \(Hx = \lambda x\).
If we multiply both sides by \(x^T\) we get \(x^THx = \lambda x^Tx\).
An eigenvector is usually scaled such that its norm is 1. The scaling doesn’t change its status of eigenvector nor the eigenvalue. This because \(H(cx) = \lambda (cx)\) where \(c\) is a scaling constant.
So if \(\|x\| = x^Tx = 1\), we get that \(x^THx = \lambda\).
Let’s verify this, before proceeding any further.
[32]:
H = np.array([[4, -1], [-1, 6]])
eigvals, eigvecs = np.linalg.eig(H)
xTHx = np.dot(eigvecs[:, 0].T, H).dot(eigvecs[:, 0])
lamxTx = eigvals[0] * np.dot(eigvecs[:, 0].T, eigvecs[:, 0].T)
assert np.isclose(xTHx, lamxTx)
assert np.isclose(xTHx, eigvals[0])
Based on the eigenvalues of \(H\) we can conclude one of the following about the curvature of a function: - the function at a point is concave up if the the eigenvalues of \(H\) are all strictly positive (positive-definite) - the function at a point is concave down if the the eigenvalues of \(H\) are all strictly negative (negative-definite) - the point of the function is a saddle point if the the eigenvalues of \(H\) are both strictly positive and negative (indefinite) - we can’t conclude anything if one of the eigenvalues is zero (positive semi-definite or negative semi-definite.)
Netwon-Raphson method with more than one variable#
Recall in the univariate case the update of the parameter was \(x_{new} = x - f'(x) / f''(x)\).
Let \(p = (x, y)\). In the bivariate case, the update of the parameter is \(p_{new} = p - H_f^{-1}(p) \cdot \nabla_f(p)\), where \(H_f^{-1}(p)\) is 2x2 matrix and \(\nabla_f(p)\) is a 2x1 column vector, so it results that \(p_{new}\) is a 2x1 column vector.
\(\begin{array}{l} \textbf{Algorithm: } \text{Newton's Method for Multivariate Optimization} \\ \textbf{Input: } \text{function to minimize } \mathcal{f}, \text{initial guess } p_0, \text{number of iterations } K, \text{convergence } \epsilon \\ \textbf{Output: } p \\ 1 : p \gets p_0 \\ 2 : \text{for k = 1 to K do} \\ 3 : \quad p_{new} = p - H^{-1}(p)\nabla f(p) \\ 4 : \quad \text{if } \|H^{-1}(p)\nabla f(p)\| < \epsilon \text{ then} \\ 5 : \quad \quad \text{return } p \\ 6 : \quad \text {end if} \\ 7 : \quad p \gets p_{new} \\ 8 : \text{return } p \\ \end{array}\)
Continuing the previous example, the gradient and the Hessian are
\(\nabla = \begin{bmatrix}4x-y\\-x+6y\end{bmatrix}\)
\(H = \begin{bmatrix}4&&-1\\-1&&6\end{bmatrix}\)
Let’s practice inverting the Hessian using the row-echelon form method.
[33]:
x, y = sp.symbols("x, y")
f = 2 * x**2 + 3 * y**2 - x * y
H = sp.hessian(f, (x, y))
HI = H.row_join(sp.Matrix.eye(2))
HI
[33]:
[34]:
HI = HI.elementary_row_op("n->n+km", row=1, k=1 / 4, row1=1, row2=0)
HI
[34]:
[35]:
HI = HI.elementary_row_op("n->kn", row=1, k=4 / 23)
HI
[35]:
[36]:
HI = HI.elementary_row_op("n->n+km", row=0, k=1, row1=0, row2=1)
HI
[36]:
[37]:
HI = HI.elementary_row_op("n->kn", row=0, k=1 / 4)
HI
[37]:
The inverse of the Hessian is
[38]:
HI[-2:, -2:]
[38]:
Let’s verify it.
[39]:
assert np.array_equal(
np.array(H.inv(), dtype="float"), np.array(HI[-2:, -2:], dtype="float")
)
Let’s calculate \(H_f^{-1} \cdot \nabla_f\)
[40]:
H.inv() @ sp.Matrix([sp.diff(f, x), sp.diff(f, y)])
[40]:
So \(\begin{bmatrix}x_{new}\\y_{new}\end{bmatrix} = \begin{bmatrix}x\\y\end{bmatrix} - \begin{bmatrix}x\\y\end{bmatrix}\)
In this case, regardless of our starting point we get that the minimum is at [0, 0] which is consistent with
\(\begin{cases}4x-y=0\\-x+6y=0\end{cases}\)
\(\begin{cases}x=\cfrac{y}{4}\\-\cfrac{y}{4}+6y=0\end{cases}\)
\(\begin{cases}x=\cfrac{y}{4}\\\cfrac{23y}{4}=0\end{cases}\)
\(\begin{cases}x=0\\y=0\end{cases}\)
Let’s verify we get the same result with the Newton’s method.
[41]:
def bivariate_newtons_method(f, symbols, initial, steps):
x, y = symbols
H = sp.hessian(f, (x, y))
hessian_eval = sp.lambdify((x, y), H, "numpy")
nabla = sp.Matrix([sp.diff(f, x), sp.diff(f, y)])
nabla_eval = sp.lambdify((x, y), nabla, "numpy")
H_dot_nabla = sp.lambdify((x, y), H.inv() @ nabla, "numpy")
p = np.zeros((steps, 2))
g = np.zeros((steps, 2))
h = np.zeros((steps, 2, 2))
step_vector = np.zeros((steps, 2))
p[0] = initial
g[0] = nabla_eval(*p[0]).squeeze()
h[0] = hessian_eval(*p[0]).squeeze()
step_vector[0] = H_dot_nabla(*p[0]).squeeze()
for i in range(1, steps):
p[i] = p[i - 1] - step_vector[i - 1]
g[i] = nabla_eval(*p[i]).squeeze()
h[i] = hessian_eval(*p[i]).squeeze()
step_vector[i] = H_dot_nabla(*p[i]).squeeze()
if np.linalg.norm(step_vector[i]) < 1e-4:
break
return p[: i + 1], g[: i + 1], h[: i + 1], step_vector[: i + 1]
x, y = sp.symbols("x, y")
f = 2 * x**2 + 3 * y**2 - x * y
p, _, h, _ = bivariate_newtons_method(
f, symbols=(x, y), initial=np.array([1, 3]), steps=10
)
display(
Math(
"\\arg \\min \\limits_{x,y}"
+ sp.latex(f)
+ "="
+ sp.latex(np.array2string(p[-1], precision=2, suppress_small=True))
)
)
print(f"Newton's method converged after {len(p)-1} steps")
print(
f"Eigenvalues of H: {np.array2string(np.linalg.eigvals(h[-1]), precision=2, suppress_small=True)}"
)
Newton's method converged after 1 steps
Eigenvalues of H: [3.59 6.41]
Since the function is quadratic Newton’s method only needed one step to find the critical point, which is a minimum because the Hessian at the critical point is positive-definite.
Let’s consider a non-quadratic function.
[42]:
x, y = sp.symbols("x, y")
g = x**4 + 0.8 * y**4 + 4 * x**2 + 2 * y**2 - x * y - 0.2 * x**2 * y
p, _, h, _ = bivariate_newtons_method(
g, symbols=(x, y), initial=np.array([4, 4]), steps=10
)
display(
Math(
"\\arg \\min \\limits_{x,y}"
+ sp.latex(g)
+ "="
+ sp.latex(np.array2string(p[-1], precision=2, suppress_small=True))
)
)
print(f"Newton's method converged after {len(p)-1} steps")
print(
f"Eigenvalues of H: {np.array2string(np.linalg.eigvals(h[-1]), precision=2, suppress_small=True)}"
)
Newton's method converged after 7 steps
Eigenvalues of H: [8.24 3.76]
The critical point \([0, 0]\) is a minimum because the Hessian is positive-definite.